PCR protocols
Outlined below are the specific reagents used and equipment utilised to optimise the various PCR reactions and what to do after obtaining strong PCR products.
All protocols were optimised at the Queensland University of Technology Molecular Genetics Research Facility.
- PCR recipes were optimised using OneTaq® 2X Hot Start Master Mix with Standard Buffer (New England Biolabs, USA).
- The recipes will be dependent on the reagents being used. The addition of BSA is necessary for the amplification of pupal cases, larvae and suboptimally preserved samples. The total amount of BSA must be reduced to 0.5 µL if PCR cycling is increased above 40 cycles, and this is recommended for degraded specimens. Addition of BSA for PCRs of fresh adult DNA is optional; however, the addition of BSA has consistently resulted in yielding brighter PCR product. BSA (10% concentration) was made up from lyophilized powder (>96%) from Sigma-aldrich CAT#A2153
- Reactions were carried out in an Eppendorf Mastercycler Pro (Eppendorf, Australia)
- PCR products were run on a 1.5% Agarose gel to assess amplification success
- Amplicons were purified using an ISOLATE PCR and Gel Kit® (Bioline)
- PCR products can be sent to external facility (e.g. Micromon, Monash University or Macrogen, Korea) for DNA sequencing in both directions using the relevant (gene specific) PCR primers (Note, depending on each laboratories procedures, and quality of the sequence obtained therein, sequencing may be conducted in one direction only).
After high quality DNA sequences have been obtained (preferably with a QV or Phred score of greater than 20), and with PCR primer sequence removed and the forward and reverse consensus sequence determined (if sequencing both directions), they can be compared with a public database (i.e. NCBI Genbank, Barcode of Life Data Systems website) to identify species.
Agarose gel electrophoresis results demonstrating amplification success for COI PCRs with and without BSA for larval and adult fruit flies
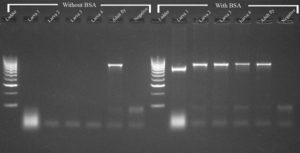
PCR products generated using Dacine specific COI primers with and without the addition of 0.5 µL BSA per reaction
Amplification of diagnostic loci
Mitochondrial cytochrome c oxidase subunit I (COI)
The DNA barcode region of the COI gene is PCR amplified using a fruit fly-specific priming system, producing a ~550 bp amplicon for sequencing.
Download method
Nuclear eukaryotic translation initiation factor 3 subunit l (EIF3L)
EIF3L is a component of the eukaryotic translation initiation factor 3 (EIF-3) complex, which is required for several steps in the initiation of protein synthesis.
Download method
Nuclear ribonuclease p protein subunit p29 (POP4)
POP4 is a part of a protein complex that generates mature tRNA molecules by cleaving their 5’ ends.
Download method
Nuclear replication protein a 32 kda subunit (RPA2)
RPA2 is part of the heterotrimeric replication protein A complex (RPA/RP-A), which binds and stabilises single-stranded DNA intermediates that form during DNA replication or upon DNA stress.
Download method
Nuclear dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit 2 isoform x2 (DDOSTs2)
Essential subunit of the N-oligosaccharyl transferase (OST) complex which catalyses critical reactions between sugars, peptides and lipids in the endoplasmic reticulum.
Download method
